Implementing Your Own k-Nearest Neighbor Algorithm Using Python
A detailed explanation of one of the most used machine learning algorithms, k-Nearest Neighbors, and its implementation from scratch in Python. Enhance your algorithmic understanding with this hands-on coding exercise.
Measuring distance between all cases
Given a new flower from an unknown species, you want to assign it a species based on what other flowers it is most similar to. For this, you need a measure of similarity. One such measure is the Euclidean distance, where distance d between two points (a1, a2) and (b1, b2) is given by d = sqrt((a1-b1)^2 + (a2-b2)^2). Each flower in the iris dataset has 4 dimensions (i.e. 4 features), and so you write a function to find the distance between each flower. The previous distance formula generalises to higher dimensions, such that the distance between two points (a1, a2, a3, a4) and (b1, b2, b3, b4) is simply
We’ll be making use of the zip function and list comprehensions. The zip function aggregates elements from lists (or other iterables, like strings) to return a list of tuples, such that zip([1,2,3], [4,5,6]) will return [(1,4), (2,5), (3,6)]. List comprehensions are a powerful Pythonic construct that facilitate quick computations on lists. For example, you could quickly get the square of all numbers 1 to 100 with [pow(x, 2) for x in range(101)], or double the values of odd numbers from 1 to 10 with [x*2 for x in range(11) if x%2==1]. Here, you are iterating over values from the corresponding dimensions in the two data points, calculating the differences squared, and storing each dimension's differences squared in diffs_squared_distance. These are then summed and returned.

For example, you can now get the distance between the first two training data instances:
Or any other tuple you'd like:
Getting the distances to all neighbours
Next, a function that returns sorted distances between a test case and all training cases is needed. One solution is the following:
Let's unpack these function: first, you need to calculate the distances from any given test data point to all the instances in the training sets. This can be done by iterating over each training_instance in training_set, and using the helper function _get_tuple_distance (the leading underscore indicates the function is intended for internal use only) to calculate the distance between it and the test instance. It also handily returns the training instance it's working on (the usefulness of this will become apparent later). To test this function, try:
This returns the train instances train[0] itself, followed by the distance (1.23) between it and the test[0] instance.
This pairwise calculation is done for every train instance and the given test instance. You can get a feel for what is happening with this command:
Next, the distances (e.g. like the 1.23, 4.47, 4.26 here) are sorted in order to find the k closest neighbours to the test instance. To understand how we can return the sorted distances, play with:
Which are the training instances ranked from closest to furthest from our test instance, as desired. The function takes the k parameter, which controls how many nearest neighbours are returned.
As a side note, instead of using a sort to order the distances from a test case in decreasing order, it would be computationally cheaper to find the maximum in the list of distances - specifically, the sort here has complexity n log(n) while passing through an array to find the maximum is O(n). If you were optimising our script for efficiency (rather than focusing on doing an educational demo), these types of considerations would become quite important.
Given a new flower from an unknown species, you want to assign it a species based on what other flowers it is most similar to. For this, you need a measure of similarity. One such measure is the Euclidean distance, where distance d between two points (a1, a2) and (b1, b2) is given by d = sqrt((a1-b1)^2 + (a2-b2)^2). Each flower in the iris dataset has 4 dimensions (i.e. 4 features), and so you write a function to find the distance between each flower. The previous distance formula generalises to higher dimensions, such that the distance between two points (a1, a2, a3, a4) and (b1, b2, b3, b4) is simply
We’ll be making use of the zip function and list comprehensions. The zip function aggregates elements from lists (or other iterables, like strings) to return a list of tuples, such that zip([1,2,3], [4,5,6]) will return [(1,4), (2,5), (3,6)]. List comprehensions are a powerful Pythonic construct that facilitate quick computations on lists. For example, you could quickly get the square of all numbers 1 to 100 with [pow(x, 2) for x in range(101)], or double the values of odd numbers from 1 to 10 with [x*2 for x in range(11) if x%2==1]. Here, you are iterating over values from the corresponding dimensions in the two data points, calculating the differences squared, and storing each dimension's differences squared in diffs_squared_distance. These are then summed and returned.
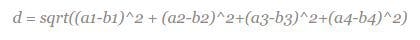
import math # 1) given two data points, calculate the euclidean distance between them def get_distance(data1, data2): points = zip(data1, data2) diffs_squared_distance = [pow(a - b, 2) for (a, b) in points] return math.sqrt(sum(diffs_squared_distance))
For example, you can now get the distance between the first two training data instances:
>>> get_distance(train[0][0], train[1][0]) 4.052159917870962
Or any other tuple you'd like:
>>> get_distance([1,1], [2,3]) 2.23606797749979
Getting the distances to all neighbours
Next, a function that returns sorted distances between a test case and all training cases is needed. One solution is the following:
from operator import itemgetter def get_neighbours(training_set, test_instance, k): distances = [_get_tuple_distance(training_instance, test_instance) for training_instance in training_set] # index 1 is the calculated distance between training_instance and test_instance sorted_distances = sorted(distances, key=itemgetter(1)) # extract only training instances sorted_training_instances = [tuple[0] for tuple in sorted_distances] # select first k elements return sorted_training_instances[:k] def _get_tuple_distance(training_instance, test_instance): return (training_instance, get_distance(test_instance, training_instance[0]))
Let's unpack these function: first, you need to calculate the distances from any given test data point to all the instances in the training sets. This can be done by iterating over each training_instance in training_set, and using the helper function _get_tuple_distance (the leading underscore indicates the function is intended for internal use only) to calculate the distance between it and the test instance. It also handily returns the training instance it's working on (the usefulness of this will become apparent later). To test this function, try:
>>> _get_tuple_distance(train[0], test[0][0]) (array([array([ 4.8, 3.4, 1.6, 0.2]), 0], dtype=object), 1.2328828005937953)
This returns the train instances train[0] itself, followed by the distance (1.23) between it and the test[0] instance.
This pairwise calculation is done for every train instance and the given test instance. You can get a feel for what is happening with this command:
>>> [_get_tuple_distance(training_instance, test[0][0]) for training_instance in train[0:3]] [(array([array([ 4.8, 3.4, 1.6, 0.2]), 0], dtype=object), 1.2328828005937953), (array([array([ 5.7, 2.5, 5. , 2. ]), 2], dtype=object), 4.465422712353221), (array([array([ 6.3, 2.7, 4.9, 1.8]), 2], dtype=object), 4.264973622427225)]
Next, the distances (e.g. like the 1.23, 4.47, 4.26 here) are sorted in order to find the k closest neighbours to the test instance. To understand how we can return the sorted distances, play with:
>>> distances = [_get_tuple_distance(training_instance, test[0][0]) for training_instance in train[0:3]] >>> sorted_distances = sorted(distances, key=itemgetter(1)) >>> [tuple[0] for tuple in sorted_distances] [array([array([ 4.8, 3.4, 1.6, 0.2]), 0], dtype=object), array([array([ 6.3, 2.7, 4.9, 1.8]), 2], dtype=object), array([array([ 5.7, 2.5, 5. , 2. ]), 2], dtype=object)]
Which are the training instances ranked from closest to furthest from our test instance, as desired. The function takes the k parameter, which controls how many nearest neighbours are returned.
As a side note, instead of using a sort to order the distances from a test case in decreasing order, it would be computationally cheaper to find the maximum in the list of distances - specifically, the sort here has complexity n log(n) while passing through an array to find the maximum is O(n). If you were optimising our script for efficiency (rather than focusing on doing an educational demo), these types of considerations would become quite important.
