Dataset Splitting Best Practices in Python
If you are splitting your dataset into training and testing data you need to keep some things in mind. This discussion of 3 best practices to keep in mind when doing so includes demonstration of how to implement these particular considerations in Python.
So you have a monolithic dataset and need to split it into training and testing data. Perhaps you are doing so for supervised machine learning and perhaps you are using Python to do so.
This is a discussion of three particular considerations to take into account when splitting your dataset, the manner in which to deal with these considerations, and how to practically implement these considerations using Python.
For our examples we will use Scikit-learn's train_test_split module, which is useful for splitting your datasets whether or not you will be using Scikit-learn to perform your machine learning tasks. You could manually perform these splits some other way (using solely Numpy, perhaps), but the Scikit-learn module includes some useful functionality to make this a bit easier. But pay attention; perhaps you have been using this module to perform your data splits in the past but have not taken certain of these considerations into account while doing so.
1. Randomly shuffling instances
The first consideration is: are your instances shuffled? So long as there is no reason for not shuffling our data (your data is time series, for example), we want to make certain that our instances are not just sequentially split as they are encountered in the dataset, as our instances may have been added in such a way that will introduce some unwanted bias into our model.
For example, look at how the version of the iris dataset included with Scikit-learn has its instances arranged upon loading:
from sklearn.datasets import load_iris iris = load_iris() X, y = iris.data, iris.target print(f"Dataset labels: {iris.target}")
Dataset labels: [0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2]
If you were to split your dataset with 3 classes of equal numbers of instances as 2/3 for training and 1/3 for testing, your newly separated datasets would have zero label crossover. That's obviously a problem when trying to learn features to predict class labels. Thankfully, the train_test_split module automatically shuffles data first by default (you can override this by setting the shuffle parameter to False).
To do so, both the feature and target vectors (X and y) must be passed to the module. You should set a random_state for reproducibility. Either train_size or test_size needs to be set, but both are not necessary. If you do explicitly set both, they must add up to 1.
from sklearn.model_selection import train_test_split X_train, X_test, y_train, y_test = train_test_split(X, y, train_size=0.67, random_state=42) print(f"Train labels:\n{y_train}") print(f"Test labels:\n{y_test}")
Train labels: [1 2 1 0 2 1 0 0 0 1 2 0 0 0 1 0 1 2 0 1 2 0 2 2 1 1 2 1 0 1 2 0 0 1 1 0 2 0 0 1 1 2 1 2 2 1 0 0 2 2 0 0 0 1 2 0 2 2 0 1 1 2 1 2 0 2 1 2 1 1 1 0 1 1 0 1 2 2 0 1 2 2 0 2 0 1 2 2 1 2 1 1 2 2 0 1 2 0 1 2] Test labels: [1 0 2 1 1 0 1 2 1 1 2 0 0 0 0 1 2 1 1 2 0 2 0 2 2 2 2 2 0 0 0 0 1 0 0 2 1 0 0 0 2 1 1 0 0 1 2 2 1 2]
You can see that we now have our instances shuffled.
2. Stratifying classes
The next consideration is this: are the class counts of our newly-split training and testing datasets evenly distributed?
import numpy as np print(f"Numbers of train instances by class: {np.bincount(y_train)}") print(f"Numbers of test instances by class: {np.bincount(y_test)}")
Numbers of train instances by class: [31 35 34] Numbers of test instances by class: [19 15 16]
This is not an even split. This relates to the idea of whether our algorithm gets equal opportunity to learn the features of each of the dataset's classes, and subsequently test what it has learned on the same number of instances of each class. This is of particular interest in smaller datasets, but should be a persistent concern.
We can force the class proportion across train and test splits with train_test_split's stratify option, noting that we will stratify with respect to the class distribution in y.
X_train, X_test, y_train, y_test = train_test_split(X, y, train_size=0.67, random_state=42, stratify=y) print(f"Train labels:\n{y_train}") print(f"Test labels:\n{y_test}")
Train labels: [2 0 2 1 0 0 0 2 0 0 1 0 1 1 2 2 0 0 2 0 2 0 0 2 0 1 2 1 0 1 0 2 1 2 1 0 2 0 2 0 1 1 0 2 1 1 0 2 1 2 0 1 0 2 1 1 1 1 1 1 2 1 2 2 0 2 1 1 2 0 2 2 2 0 2 0 0 2 2 2 0 1 2 2 0 1 1 1 1 1 0 2 1 2 0 0 1 0 1 0] Test labels: [2 1 0 1 2 1 1 0 1 1 0 0 0 0 0 2 2 1 2 1 2 1 0 2 0 2 2 0 0 2 2 2 0 1 0 0 2 1 1 1 1 1 0 0 2 1 2 2 1 2]
print(f"Numbers of train instances by class: {np.bincount(y_train)}") print(f"Numbers of test instances by class: {np.bincount(y_test)}")
Numbers of train instances by class: [34 33 33] Numbers of test instances by class: [16 17 17]
This looks better now, and the raw numbers tell us that this is the most optimally stratified split possible.
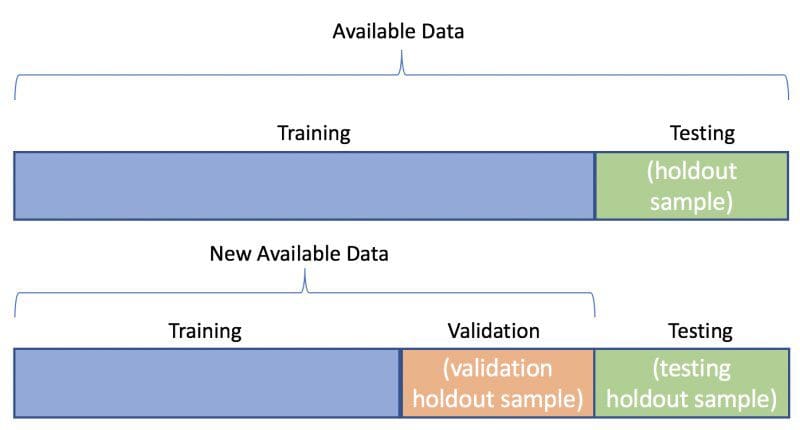
3. Splitting the splits
The third consideration relates to our testing data: is our modeling task content having only a single testing dataset, made up of previously-unseen data, or should we be using two such sets — one for validating our model during its fine-tuning, and perhaps multiple models, and another as a final holdout set for model comparison and selection.
If we chose 2 such sets, this would mean that there would be one set of data that is held out until all assumptions have been tested, all hyperparameters tuned, and all models trained to their best performance, which is then only seen once by the models as the final step in our experiments. There is a body of work you can consult on testing versus validation sets; I won't go any further into the reasoning and arguments here.
Assuming, however, that you conclude you do want to use testing and validation sets (and you should conclude this), crafting them using train_test_split is easy; we split the entire dataset once, separating the training from the remaining data, and then again to split the remaining data into testing and validation sets.
Below, using the digits dataset, we split 70% for the training dataset, and temporarily assign the remainder to the testing set. We continue to enforce the best practices covered above.
from sklearn.datasets import load_digits digits = load_digits() X, y = digits.data, digits.target X_train, X_test, y_train, y_test = train_test_split(X, y, train_size=0.7, random_state=42, stratify=y) print(f"Numbers of train instances by class: {np.bincount(y_train)}") print(f"Numbers of test instances by class: {np.bincount(y_test)}")
Numbers of train instances by class: [124 127 124 128 127 127 127 125 122 126] Numbers of test instances by class: [54 55 53 55 54 55 54 54 52 54]
Note the stratified classes across the training and temporary testing sets.
We then re-split the testing set in the same way — this time modifying the output variable names, the input variable names, and being careful to change the stratify class vector reference — using a 50/50 split for the testing and validation sets.
X_test, X_val, y_test, y_val = train_test_split(X_test, y_test, train_size=0.5, random_state=42, stratify=y_test) print(f"Numbers of test instances by class: {np.bincount(y_test)}") print(f"Numbers of validation instances by class: {np.bincount(y_val)}")
Numbers of test instances by class: [27 27 27 27 27 28 27 27 26 27] Numbers of validation instances by class: [27 28 26 28 27 27 27 27 26 27]
Note, again, the class stratification across all datasets, which is optimal and ideal.
You are now ready to train, validate, and test as many machine learning models as you see fit with your data.
Another consideration: you might want to think about using cross-validation instead of a simple train/test or train/validate/test strategy. We will cover cross-stratification considerations next time.
Related:
- Sparse Matrix Representation in Python
- 5 Great New Features in Scikit-learn 0.23
- The 5 Most Useful Techniques to Handle Imbalanced Datasets