Analyzing Scientific Articles with fine-tuned SciBERT NER Model and Neo4j
In this article, we will be analyzing a dataset of scientific abstracts using the Neo4j Graph database and a fine-tuned SciBERT model.
By Khaled Adrani, UBIAI
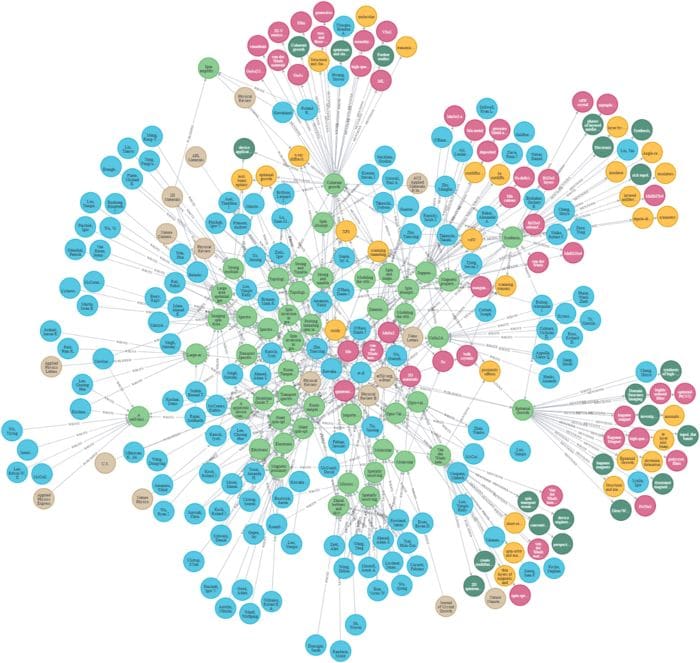
Image by Author: Knowledge graph of scientific articles
It is estimated that 1.8 million articles are published each year, in about 28,000 journals. The publications throughput has increased by 4% each since the last decade and grew from 1.8 million to 2.6 million from 2008 to 2018. But who actually read those papers? According to one 2007 study, not many people: half of the academic papers are read-only by their authors and journal editors. Analyzing articles manually is tedious and time-consuming. Therefore providing researchers with the tool that quickly extracts and analyze information from articles will have a tremendous impact on accelerating new discoveries.
Knowledge graphs KG present an ideal solution for quick and rapid analysis of information. They represent a network of real-world entities like objects and concepts and provide the relationships between them. This information is usually stored in a graph database and visualized as a graph structure. However, building a knowledge graph manually is a time-consuming task. Fortunately, with the recent advancement in Machine Learning and Natural Language Processing, Named Entity Recognition (NER) came to the rescue.
In this article, we will be analyzing a dataset of scientific abstracts using the Neo4j Graph database and a fine-tuned SciBERT model. After, we will be querying the data to answer some questions as our analysis for this corpus. This article assumes that you have the basic fundamentals of NLP and Neo4j.
Model training
The NER model we will be using is based on SciBERT and has been fine-tuned on scientific articles by annotating Materials, processes, and tasks:
- Material: Represents any mention of material in the abstract
- Process: Represents a process or a method used in the experiment
- Task: Represents the task of the study to be carried
For the annotation part, we have used UBIAI text annotation tool and have exported the annotation in IOB format as shown below:
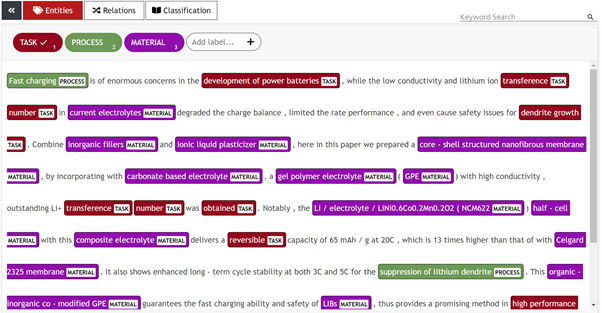
Image by Author: UBIAI Text Annotation Tool
For more information on how to generate training data using UBIAI and fine-tuning the NER model, check out the articles below:
- Introducing UBIAI: Easy-to-Use Text Annotation for NLP Applications
- How to Train a Joint Entities and Relation Extraction Classifier using BERT Transformer with spaCy 3
- How to Fine-Tune BERT Transformer with spaCy 3
Setup
We will be working on Google Collaboratory. Obviously, we will be using Python. We mount our google drive which contains the dataset and the model. We need also to install various dependencies.
#Mount google drive from google.colab import drive drive.mount(‘/content/drive’) !pip install neo4j !pip install -U spacy !pip install -U pip setuptools wheel !python -m spacy download en_core_web_trf !wget https://developer.nvidia.com/compute/cuda/9.2/Prod/local_installers/cuda-repo-ubuntu1604-9-2-local_9.2.88-1_amd64 -O cuda-repo-ubuntu1604-9-2-local_9.2.88-1_amd64.deb !dpkg -i cuda-repo-ubuntu1604-9-2-local_9.2.88-1_amd64.deb !apt-key add /var/cuda-repo-9-2-local/7fa2af80.pub !apt-get update !apt-get install cuda-9.2 !pip install torch==1.7.1+cu92 torchvision==0.8.2+cu92 torchaudio==0.7.2 -f https://download.pytorch.org/whl/torch_stable.html !pip install -U spacy[cuda92,transformers]
Let us load our NER model:
import spacy
nlp = spacy.load("/content/drive/MyDrive/Public/model_science/model-best")
Data Preparation
The dataset contains the abstracts of scientific articles written by the main authors and published in various scientific journals. We are also interested in the entities mentioned within these articles. So, you can see clearly how our graph will be structured. Let us load our data and get a look at some examples:
import pandas as pd
path = "/content/drive/MyDrive/Public/Database_ABI_updated.csv"
df = pd.read_csv(path)[["Title","Authors","Journal","DOI","Abstract"]]
df.head()

A sample of our dataset
To make our knowledge graph, we will add the authors, the journals, and the articles with their properties, then we will add existing relationships between them.
We start easy by extracting the list of authors. Some preprocessing was required. Each article usually has multiple authors and so for each article, we transformed the string containing their names into a list. We will also need to produce ids through hashing our entities.
import hashlib
ls = list(df.Authors)
ls_authors = []
for e in ls:
sep = ";"
if ";" in e:
sep = ";"
else:
sep = ","
ls_authors.extend(e.split(sep))
print(len(ls_authors))
ls_authors = list(set(ls_authors))
print(len(ls_authors))
def hash_text(text):
return hashlib.sha256(str(text).encode('utf-8')).hexdigest()
authors = []
for e in ls_authors:
authors.append({"name":e,"id":hash_text(e)})
Here is the code to get the list of all journals:
journals = []
for j in list(df.Journal.unique()):
journals.append({"name":j,"id":hash_text(j)})
journals[0]
For the articles, we will be transforming the dataframe into a list of dictionaries. Each article will hold the attribute of its own dictionary as properties in the graph (like name, list of authors, etc).
import copy
records = df.to_dict("records")
def extract_authors(text):
ls_authors = []
sep = ";"
if ";" in text:
sep = ";"
else:
sep = ","
ls_authors.extend(text.split(sep))
return ls_authors
articles = copy.deepcopy(records)
for r in articles:
r["Authors"] = extract_authors(r['Authors'])
To extract entities from each article, we will combine its title and its abstract as the text to be analyzed. At the same time, we added its own id by making it the hash of its text content:
for article in articles:
article["text"]= article["Title"]+" "+article["Abstract"]
article["id"] = hash_text(article["text"])
article
Now, this function served me well as a reusable code to work with a list of documents. The labels of our entities are:
- PROCESS
- MATERIAL
- TASK
def extract_ents(articles,nlp):
texts = []
for article in articles:
texts.append(article["text"])
docs = list()
for doc in nlp.pipe(texts, disable=["tagger", "parser"]):
dictionary=dict.fromkeys(["text", "annotations"])
dictionary["text"]= str(doc)
dictionary['id'] = hash_text(dictionary["text"])
annotations=[]
for e in doc.ents:
ent_id = hash_text(e.text)
ent = {"start":e.start_char,"end":e.end_char, "label":e.label_.upper(),"text":e.text,"id":ent_id}
annotations.append(ent)
dictionary["annotations"] = annotations
docs.append(dictionary)
return docs
docs = extract_ents(articles,nlp)
Let us see what are the entities extracted from the first article:
for e in docs[0]['annotations']:
print(e['text'],' --> ',e['label'])
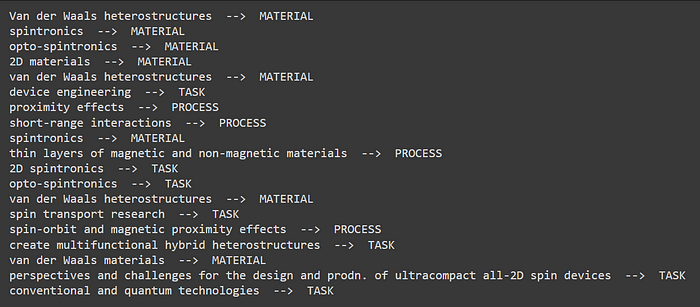
An example of entities present within an article
Finally, we add every dictionary to its appropriate article dictionary in the articles list:
for i in range(len(docs)):
articles[i]['annotations'] = copy.deepcopy(docs[i]['annotations'])
Creating the Knowledge Graph
Data preparation is done. Now is the time to insert and manipulate it using Neo4j!
We defined a function that we will be using to communicate with our instance from the Neo4j Aura database.
from neo4j import GraphDatabase
import pandas as pd
uri ="your uri here"
user="your username here"
password='your password here'
driver = GraphDatabase.driver(uri,auth=(user, password))
def neo4j_query(query, params=None):
with driver.session() as session:
result = session.run(query, params)
return pd.DataFrame([r.values() for r in result], columns=result.keys())
These are the queries we used to populate our database, they are very straightforward.
#create journals
neo4j_query("""
UNWIND $data as journal
MERGE (j:JOURNAL {id:journal.id})
SET j.name = journal.name
RETURN count(j)
""",{"data":journals})
#create authors
neo4j_query("""
UNWIND $data as author
MERGE (a:AUTHOR {id:author.id})
SET a.name = author.name
RETURN count(a)
""",{"data":authors})
#create articles
neo4j_query("""
UNWIND $data as row
MERGE (a:ARTICLE{id:row.id})
ON CREATE SET a.title = row.Title, a.DOI = row.DOI, a.abstract = row.Abstract,
a.authors = row.Authors, a.journal=row.Journal
RETURN count(*)
""", {'data': articles})
# Match articles with their authors
neo4j_query("""
MATCH (a:ARTICLE)
WITH a
UNWIND a.authors as name
MATCH (author:AUTHOR) where author.name = name
MERGE (author)-[:WROTE]->(a)
""")
# Match articles with their journals
neo4j_query("""
MATCH (a:ARTICLE)
WITH a
MATCH (j:JOURNAL) where j.name = a.journal
MERGE (j)-[:PUBLISHED]->(a)
""")
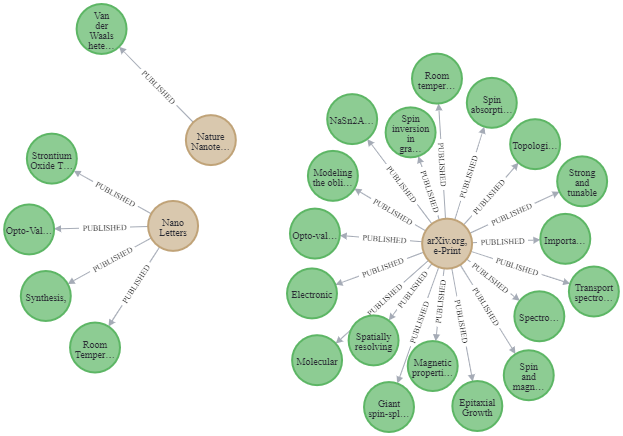
Image by Author: Three journals and their published articles
Adding the entities is a little bit tricky. This query is composed of three parts:
- First, we match every article node in the database with its own dictionary from our list of articles. UNWIND helps us loop through the list and get one article each time.
- Second, for every entity, we create it if it does not exist or match it by using MERGE. After, the entity we will be linked with the current article.
- Last, we add the label PROCESS, MATERIAL, or TASK for every entity, the value of the label is already contained within a property with the same name. Then we proceed to remove that property.
# Add entities (Material, Process, Task) and match them with any article that mentions them.
neo4j_query("""
UNWIND $data as row
MATCH (a:ARTICLE) where row.id = a.id
WITH a, row.annotations as entities
UNWIND entities as entity
MERGE (e:ENTITY {id:entity.id})
ON CREATE SET
e.name = entity.text,
e.label = entity.label
MERGE (a)-[m:MENTIONS]->(e)
ON CREATE SET m.count = 1
ON MATCH SET m.count = m.count + 1
WITH e as e
CALL apoc.create.addLabels( id(e), [ e.label ] )
YIELD node
REMOVE node.label
RETURN node
""", {'data': articles})
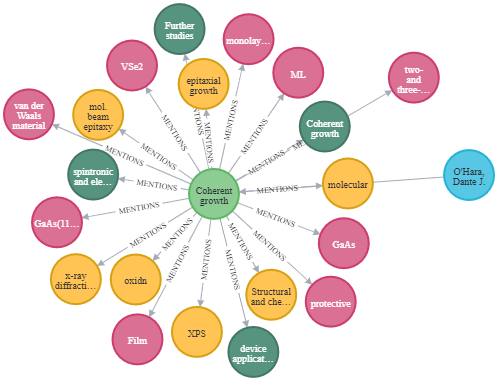
Image by Author: An article and its connected nodes
Abstract Analysis
Finally, we arrive at the most interesting part!
Say that you are an NLP expert working with a physics expert. He wants you to analyze the abstracts of some scientific papers he finds very interesting. He gives you four questions:
1) Most popular material and process
2) Most popular author
3) Highest co-occurrence between materials and processes
4) Shortest path between two given authors
Let us answer these questions using Neo4j!
To find out the most popular material and process in the entirety of our corpus, we need to count how many relations one entity has with journals, to see how many times they appeared:
neo4j_query("""
MATCH (e) where e:PROCESS OR e:MATERIAL
MATCH (e)-[]-(a:ARTICLE)-[]-(j:JOURNAL)
RETURN e.name as entity, labels(e) as label, count(*) as freq ORDER by freq DESC LIMIT 10
""")

Top ten popular materials and processes in our corpus
The most popular author can be obtained using the same reasoning and here is the query for that:
neo4j_query("""
MATCH (a:AUTHOR)-[]-(ar:ARTICLE)-[]-(j:JOURNAL)
RETURN a.name as author, count(*) as freq
ORDER BY freq DESC
LIMIT 10
""")
Now, Co-occurrence analysis is the counting of the occurrences of a pair of entities within the document. For example, we want to know how many times a certain process and a certain material were mentioned together within the same article.
neo4j_query("""
MATCH (m:MATERIAL)<-[:MENTIONS]-(a:ARTICLE)-[:MENTIONS]->(p:PROCESS)
WHERE id(m) < id(p)
RETURN m.name as MATERIAL, p.name as PROCESS, count(*) as cooccurrence
ORDER BY cooccurrence
DESC LIMIT 5
""")
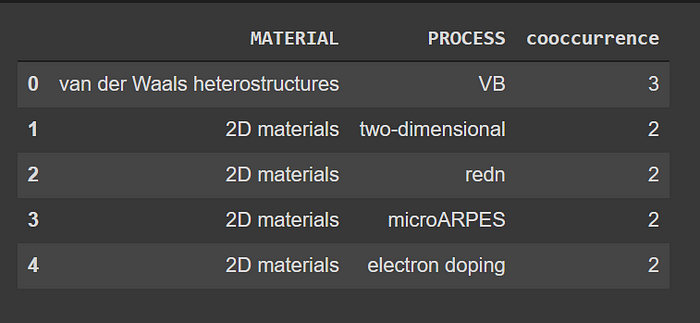
Co-occurrence analysis output
We can see that the material van der Waals heterostructures and the term VB were mentioned three times together in the entirety of the corpus. VB is the acronym of Valence Band which is the highest energy an electron can jump out of, moving into the conduction band when excited and it plays a very important role in van der Waals structures. Using our knowledge graph we have discovered this correlation semantically without any prior knowledge! Such analysis can help us find new connections and unseen correlations between scientific concepts.
Lastly, we want to find the shortest path between two given authors, for that, we match each one with their respective id, use the predefined function shortestPath which accepts a path as an input, and then we get our result. We ran this query directly on the Neo4j browser to be able to get the graph picture.
MATCH (a1:AUTHOR ),
(a2:AUTHOR ),
p = shortestPath((a1)-[*]-(a2))
where a1.id = '6a2552ac2861474da7da6ace1240b92509f56a6ec894d3e166b3475af81e65ae' AND a2.id='3b7d8b78fc7b097e2fd29c2f12df0abc90537971e2ac5c774d677bcdf384a3b7'
RETURN p

Image by Author: Shortest path between two given authors
Interestingly enough, we can predict that these two authors might need a common article for their research, or that they can actually cooperate together in some form or another. Visualizing meaningful relationships is of utmost importance in making informed decisions.
Conclusion
This was a demonstration of the power of combining Named Entity Recognition and Knowledge Graph in Text Mining. We did not delve into details because we wanted to focus more on showcasing the workflow of handling text semantic analysis. I hope that you have learned a thing or two and in the future, we will be delving more in-depth!
If you have any questions or want to create custom models for your specific case, leave a note below or send us an email at admin@ubiai.tools.
Follow us on Twitter @UBIAI5
Bio: Khaled Adrani is a Computer Science Engineer and intern at UBIAI.
Original. Reposted with permission.
Related:
- Build a Serverless News Data Pipeline using ML on AWS Cloud
- Meta-Learning for Keyphrase Extraction
- The Ultimate Guide To Different Word Embedding Techniques In NLP
