Simple Ways Of Working With Medium To Big Data Locally
An overview of the installation and implementation of simple techniques for working with large datasets in your machine.
By Francisco Juretig, creator of nitroproc
As data scientists, we are typically exposed to datasets ranging from a few dozen million to hundreds of millions of records, with possibly dozens or hundreds of columns. This data is typically received in csv files, and processed in many ways with the ultimate intention of running a complicated ML algorithm in either Python or R. However, this data will not fit into the RAM memory, so alternative tools need to be devised. Naturally, all this data could be allocated in a cluster and processed there; but it requires a cumbersome process of uploading files, paying for server time, security concerns if we are dealing with sensitive data, quite sophisticated programming techniques, and spending time downloading the outputs. Of course, a local Spark session can always be created, but the learning curve is substantial and installing it might be challenging to beginners (especially if the objective is to process a few big files occasionally). In this article, we focus on simple techniques (both for installing and running) for working with this kind of datasets.
First option: Looping through the files
Python or R can always be used to quickly loop through files. In this case we will use to loop through a very big file. The dataset is the following: 5,379,074 observations and 8 columns, with integers, dates and strings, with a file size of 212Megabytes on disc.

We will generate a class for working with this file. In this class we will define two simple operations, the first one will calculate the maximum, and the second one will filter the records corresponding to a particular name. In the last three lines, we instantiate this class, and consume its methods. Notice that we are not using pandas.read_csv since this dataset will generally not fit into memory (depending on your computer). Also notice we are excluding the first record, since it contains the file headers.

The problem is that in most practical scenarios, the very same moment we deviate from simple sub-setting operations, the complexity of the code will increase in an astonishing way. For example, even coding a simple sort procedure using files is incredibly complicated, let alone working with missing values, multiple variable types, date formats, etc. What’s worse is that most data processing operations will require a good implementation of a sort procedure: merging, transposing, and summarizing are not only difficult by themselves, but they will always require sorted data to run on reasonable time.
Second option: SAS
SAS has been the best software for statistical analysis for many decades. Apart from its excellent statistical methods implementations, SAS allows us to work with hundreds of millions of observations easily. It achieves this trick by using the hard drive to store the data, instead of the RAM memory (as most software would do). Apart from that, these files can have thousands of columns. The SAS programming language is very easy and flexible. For instance, reading a file and sorting it can be achieved in very few lines:
Data customer_data;
infile “./sales.csv” lrecl=32767 missover;
input User_id: 8. Salesman: $10. Date: mmddyy8. Customername: $10. City:$3. Country: $6.
Discount: $9. Promo: 8.;
run;
proc sort data = customer_data;
by User_id;
run;
A usually overlooked aspect of SAS, yet one of its strongest features, are its log files. They are an extremely powerful tool for identifying weird observations, missing values, abnormal results. For example, when merging two files with millions of observations, you can immediately identify problems by looking into how many observations where read in both files versus how much was written as an output.
You won’t generally find any sort of restriction or limitation. Yet for extremely big files, it will be more convenient to work with a distributed environment, using Spark running on a cluster (but that is beyond the scope of this article). Unfortunately, SAS licenses will generally be quite expensive, and it only runs on computers.
Third option: nitroproc
nitroproc is a free cross platform software (currently available in Windows/MacOS/Android/iOS) designed for data manipulation (specially for data scientists). It can be called via batch mode through Python or R. In a similar fashion to SAS, it is designed to operate using the hard drive, so there are practically no limits on the data size. Its scripts can be deployed to any device, with no changes to its syntax (except for obviously the correct input file paths that will be processed). In this article, we will show how to use the Windows and Android versions (the iOS version can also be downloaded from the App Store). It handles different variable types, file formats, and missing values. The current version allows you to sort, merge, filter, subset, compute dummies, aggregate and many other data manipulation operations traditionally used by data scientists. Similarly to SAS, nitroproc produces very powerful logs for identifying weird data, wrong merges, etc. Additionally, it also produces another log file, called logtracer used for logically analysing how the different instructions in your script are related.
Sorting 5.4 million observations by one key
In this example, we will sort a very big file in both a PC and an (old) Android phone, just to show the sheer power of nitroproc. We will use the same dataset we used in Figure 1 5.4 million records dataset. Remember that this dataset has 5,379,074 observations and 8 columns, with integers, dates and strings, with a file size of 212Megabytes on disc. In this case we will sort it by its first column (User_id).
You can download the csv file from: https://www.nitroproc.com/download.html to replicate the same results we show here. In that case I suggest you run the tests with the phone connected to the PC/Mac, and with no other processes running on the background.
The syntax is really simple, we just write:
sort(file=sales.csv,by=[User_id],coltypes=[int,string,dd/mm/yyyy,string,string,string,int,int], order=[asc], outname = result.csv, out_first_row = true)
All the parameters are pretty self explanatory. Order specifies whether we want asc, or desc, out_first_row is used to specify whether we want to output the file headers or not. You might notice that we are not specifying any headers because the csv already contains them. If they were not included, we would need to type headers=[colum_name1,…,column_namek]. For PCs we need to specify proper file paths, but for the Android and iOS versions, only the file names are necessary, as the files paths are recovered automatically (for Android the /Downloads folder is used and for iPhone/iPad the App folder is used – it can be accessed via iTunes).
Sorting is particularly important in nitroproc as it used for merging files, summarizing files, and other operations.
PC Version
It takes 1 min and 25 seconds (Check Figure 3: log file produced by nitroproc - PC Version) to finish on a quite standard (from 2012) desktop Intel i5-4430 @3.00 GHz and a standard Seagate 500GB ST500DM002 hard drive. On newest Intel devices, for example i7-4970k, and using solid state drives, the scripts will be at least 3x faster (even greater speeds can be achieved via overclocking).

Android Version
Running the same script on aNexus 5x at Android 7.0 Nougat is much slower (Figure 4 log file produced by nitroproc - Android version, but it still works flawlessly). This phone was released on 2015, with a 1.8Ghz processor (bear in mind this isnot a high-end phone). As you can see in Fig1, it takes 15minutes. On newest (2017) high end Android phones, you should expect to run this script in half this time (7 minutes). Since basically no RAM is used, nitroproc can run very well in RAM deprived systems.
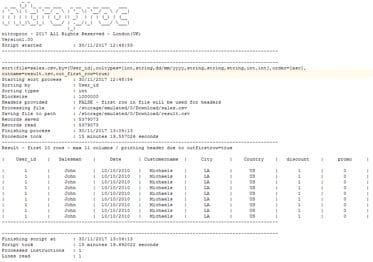
iOS Version
Finally, take a look at the output produced by the iOS version, running on an iPhone 8 Plus (A11 Bionic Chip). These results surprised me like no benchmark before (and I used to be an enthusiast overclocker). Apple claims that the iPhone 8 and X are the fastest phones ever, but that is probably an understatement. You can see that it sorts the file in 2 minutes 42 seconds, less than twice what our desktop Intel CPU takes (keep in mind a PC consumes more than 250 watts, and an iPhone 8s around 6.96 watts) and almost 5.5x faster than a regular Android phone. A more accurate statement for the iPhone 8 Plus (and iPhone X since they share the same chip) is that it is a marvel of engineering, providing a comparable performance to a desktop gaming CPU consuming 35x more power. Consider that nitroproc is very intensive in I/O operations, and in this case, there are hundreds of millions of writes and reads, since there are lots of intermediate operations. The fact that the A11, together with its operating system can move so much data so fast from its hard drive, is almost surreal.

Related
