 Python for data analysis… is it really that simple?!?
Python for data analysis… is it really that simple?!?
The article addresses a simple data analytics problem, comparing a Python and Pandas solution to an R solution (using plyr, dplyr, and data.table), as well as kdb+ and BigQuery solutions. Performance improvement tricks for these solutions are then covered, as are parallel/cluster computing approaches and their limitations.
By Ferenc Bodon Ph.D., Data Engineer, Cloud Solutions Architect at Kx

Python is a popular programming language that is easy to learn, efficient and enjoys the support of a large and active community. It is a general-purpose language with libraries specialized for various areas, including web development, scripting, data science, and DevOps.
Its primary data analysis library, Pandas, gained popularity among data scientists and data engineers. It follows Python’s principles, so it seems to be easy to learn, read and allows rapid development… at least based on the textbook examples. But, what happens if we leave the safe and convenient world of the textbook examples? Is Pandas still an easy-to-use data analysis tool to query tabular data? How does it perform compared to other professional tools like R and kdb+?
In this article, I will take an example that goes just one step beyond the simplest use cases by performing some aggregation based on multiple columns. The complexity of my use case is around level 2 out of 5 levels. Anybody who analyzes data tables will bump into the problem, probably in the second week. For comparison, I will also cover other popular tools that aim for data analysis.
- First of all, the problem can be solved by ANSI SQL so all traditional RDBM systems like PostegreSQL, MySQL, etc can enter the game. In the experiments, I will use BigQuery, a serverless, highly-scalable data warehouse solution by Google.
- The R programming language is designed for statistical analysis. It natively supports tables via its class data.frame. Using multiple aggregations is quite inconvenient due to the limitation of the core function aggregate. The R community developed the library plyr to simplify the usage of data.frame. Package plyr was retired and package dplyr was introduced with the promise of improved API and faster execution. Package dplyr is part of the tidyverse collection that is designed for professional data science work. It provides an abstract query layer and decouples the query from the data storage let it be data.frame or in an external database that supports ANSI SQL. Package data.table is an alternative of dplyr and it is famous for its speed and concise syntax. A data.table can also be queried by the dplyr syntax.
- In the Q/Kdb+ programming language, tables are also first-class citizens and the speed was a primary design concept of the language. Kdb+ made use of multicore processors and employs map-reduce from its birth in 2004 if data was partitioned on the disk. From version 4.0 (released in March 2020) most primitives (such as sum, avg, dev) use slave threads and are executed in parallel even if the table is in memory. Productivity was the other design consideration - any redundant programming element that does not contribute to the understanding (even a parenthesis) is regarded as visual noise. Kdb+ is a good contender for any data analysis tool.
I will consider the elegance, simplicity and the speed of the various solutions. Also, I investigate how to tune the performance and leverage multicore processors or cluster of computers by employing parallel computation.
The Problem
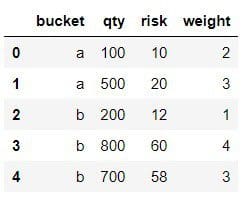
We are given a simple table with four columns, one nominal, called bucket and three numerical, qty, risk, and weight. For simplicity let us assume that the numerical columns contain integers.
We would like to see for each bucket
- the number of elements, as column NR
- the sum and the average of qty and risk, as columns TOTAL_QTY/TOTAL_RISK and AVG_QTY/AVG_RISK
- the weighted average of qty and risk, as columns W_AVG_QTY and W_AVG_RISK. Weights are provided in column weight.
To get the solution, I will not use any deprecated approach e.g renaming aggregation by a nested dictionary. Let us solve each task separately.
Number of elements in each bucket
Getting the number of elements in each bucket does not look enticing and requires intense typing

The literal bucket is required three times and you need to use 5 brackets/parentheses ????.
The solutions in R look more tempting.

Developers of libraries dplyr and data.table also have aversion to word repetition. They introduced special built-in variables n() and .N respectively that hold the number of observations in the current group. This simplifies the expressions and we can get rid of a pair of parentheses.

The ANSI SQL expression is simple and easy to understand.

You can avoid the repetition of the literal bucket by employing column indices in the GROUP BY clause. IMHO this is not a recommended design because the expression is not self-documenting and less robust. In fact, Apache deprecated usage of numbers in GROUP BY clauses.
The kdb+ expression is more elegant. It requires no brackets, quote marks or any word repetition.

SQL forms the basis of data analysis so probably everybody understands the ANSI SQL and kdb+ solutions. R and kdb+ developers agree that "GROUP BY" is too verbose, a simple "by" literal is expressive enough.
Note that apart from Pandas, no languages use any quotation marks in this simple expression. The query in Pandas requires four pairs of them ???? to wrap column names. In R, SQL, and kdb+ you can refer to columns as if they were variables. The notation .() in R - which is an alias for list() - allows this convenience feature.
Aggregation of multiple columns
Calculating sum and average of a single column and calculating the sums of multiple columns are quite simple with Pandas




Code gets nasty if you try to combine the two approaches as it results in a column name conflict. Multi-level columns and function map need to be introduced.


The SQL, R, and kdb+ equivalents do not require introducing any new concept. The new aggregations are simply separated by commas. You can use keyword sum and avg/mean to get sum and average respectively.

Observe the lightness of the kdb+ expression; it does not require parentheses or brackets.

Weighted average
The weighted average is supported by Numpy library that Pandas relies on. Unfortunately, it cannot be used in the same way such as np.sum. You need to wrap it in a lambda expression, introduce a local variable, use apply instead of agg and create a data frame from a series.


Neither standard SQL nor its Google extension, BigQuery, provides a built-in function to get a weighted average. You need to recall the definition and implement it by hand.

Again the R and Q/Kdb+ solutions do not require introducing any new concept. The function weighted average is supported natively and accepts two column names as parameters.

In kdb+ you can use infix notation to get a more natural syntax - just pronounce this "w weighted average x".

All in one statement
Let us put all the parts together. We created multiple data frames, so we need to join them


Note that the first join expression has nothing to do with the others. It creates a string from a list of strings as opposed to the others which perform left joins.
To get the final result, we need three expressions and a temporary variable res. If we spend more time searching forums then we can find out that this complexity is partially attributed to deprecating nested dictionaries in function agg. Also, we might discover an alternative, less documented approach using just the function apply, with no join. Unfortunately, this solution returns all numeric columns of type float so you need to explicitly cast integer columns.
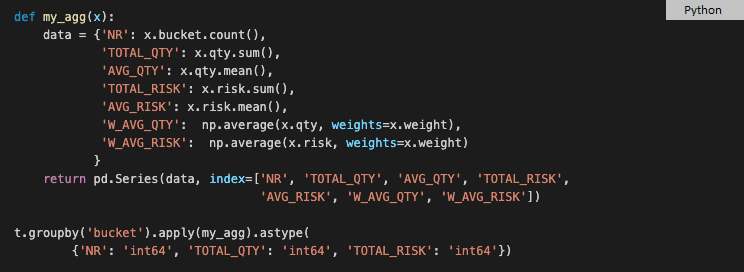
This solution requires creating a temporary function that will probably never be used again in your source code. We can squeeze all the statements into a single, stateless solution but that results in a hard-to-read and hard-to-maintain, nested code. Also, this second approach is slower on mid-size tables.
The Pandas release of the 18th of July, 2019 supported group-by aggregation by named aggregator. It provides a more elegant syntax than the aforementioned apply-based solution and does not require typecasting. Also, the developers probably recognized the pain caused by the overwhelming usage of the quotation marks. Unfortunately, the weighted average is not supported because only a single column can be used in the aggregation. For completeness, we provide the new proper syntax, ignoring the weighted average calculation. It is great to see that the output column names no longer require quotation marks.

In contrast, SQL could already provide an elegant solution 30 years ago.
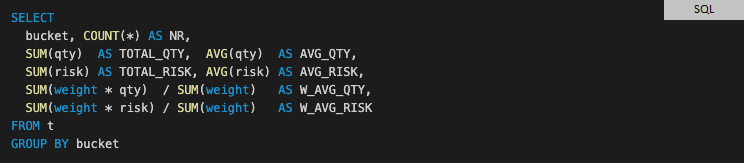
Let us see how R solves the task with a data.table

and how the solution in kdb+ appears
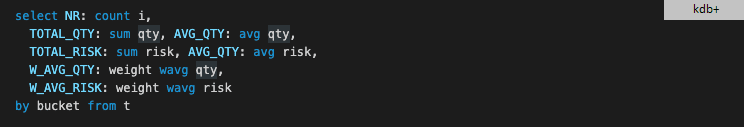
It seems that kdb+ has the most intuitive, simplest and most readable solution. It is stateless, requires no parenthesis/bracket and creation of temporary variables (or functions).
What about the performance?
The experiments were conducted on both Windows and Linux using the stable latest binaries and libraries. Queries were executed a hundred times, test Jupyter notebooks are available on Github. The data were randomly generated. The bucket fields were strings of size two and fields qty and risk are represented by 64-bit integers. Library bit64 was used in R to get 64-bit integers. The table below summarises execution times in milliseconds. The axes are log scaled. The two Python solutions of version 3.6.8 with Pandas 0.25.3 are compared to the three R libraries (version 3.6.0) and to the two kdb+ versions.
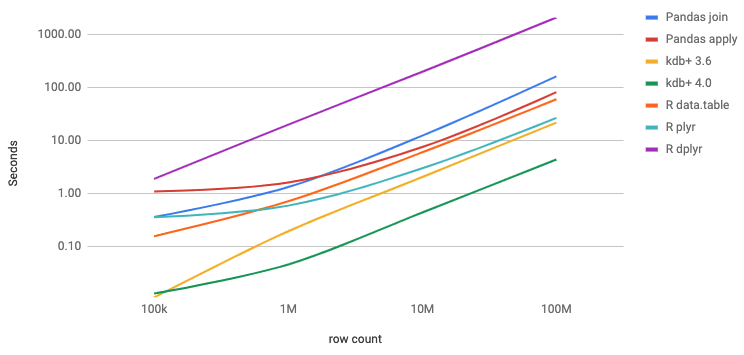
The kdb+ solution is not only more elegant than Pandas but it is also faster by more than an order of magnitude. R's data.table of version 1.12.6 is three times slower than kdb+ 3.6 for this particular query. Kdb+ 4.0 is five times faster than kdb+ 3.6 for tables of billion rows. Package dplyr of version 0.8.3 was two orders of magnitude slower than plyr of version 1.8.5 ????.
Pandas hit memory limit with an input table of 1 billion rows. All other solutions could handle that volume without running out of memory.
Let see how can we decrease execution times.
Performance optimizations
The column bucket contains strings. If the domain size is small and there are many repetitions then it is suggested to use categorical values instead of strings. Categories are like enums and represented by integers, hence they consume less memory and comparison is faster. You can convert string to categorical in Pandas by

but it is more memory efficient to create the category column during the table construct. We use function product to generate the universe of strings of length two by employing the cartesian product. In the code snippet below we omit the syntax for creating other columns. N stores the number of rows to be inserted.

Categories are called factors in R. Class data.frame converts strings to factors automatically (use "stringAsFactors = FALSE" to avoid this) but in data.table strings are left intact for good reasons.

BigQuery has no concept of category or factor. Instead, it applies various encoding and compression techniques to achieve the best performance. To generate random strings you can use Unicode code points, functions RAND and CODE_POINTS_TO_STRING and some casting. Lowercase letter "a" has code points 97 - you can figure this out by using function TO_CODE_POINTS.

You can employ a user-defined function to avoid code duplication.
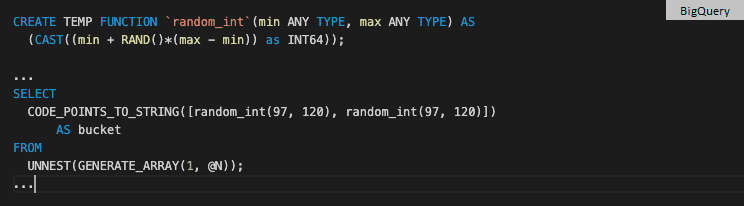
For comparison, the same operation in kdb+ looks like this

The construct N?M generates N random values depending on the type of M. If M is an integer, float, date or boolean then a random integer, float, date or boolean vector is returned. If M is a list then random list elements are picked. If N is a negative integer then the result will not contain any duplicates. In kdb+ many operators are overloaded in a similar way.
Enums are called symbols in kdb+ parlance and are heavily used by developers. The language strongly supports symbols. You can use them without defining possible values upfront, kdb+ maintains the mapping for you.
Based on my measurement, optimization by categories/symbols reduces run times by a factor of two in Pandas and kdb+. R's data.table shows different characteristics. Using factors instead of strings has no impact on the performance. This is due to the built-in string optimization via the global string pool. Although, factors are stored as 32-bit integers and strings require 64-bit pointers to the pool elements, the difference has a marginal impact on the execution times.
We can further improve performance if we use types that require less space. For example, if column qty fits into 2 bytes then we can use int16 in Pandas and short in kdb+. This results in less memory operation which is often the bottleneck in data analysis. R does not support 2-bytes integers so we used its default integer type which occupies 4 bytes.
Integers of size 32, which is the default in R, results in a 5-6 fold execution time improvements. Use 64-bit integers only if you really need the large cardinality. This is especially true for package dplyr.
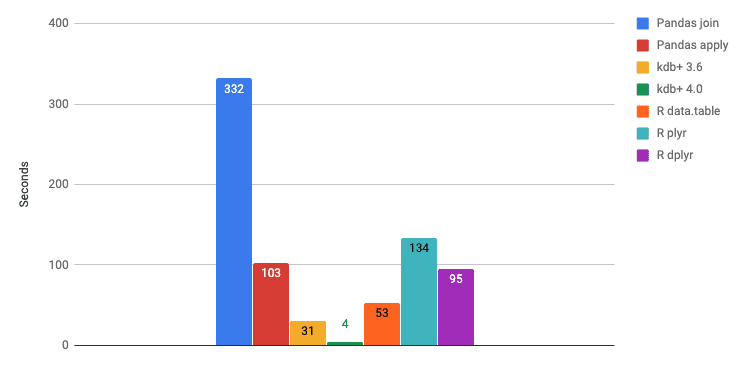
When calculating the aggregation we need to keep track of the bucket due to the grouping. If the table is sorted by the group column then aggregation is faster as groups are already gathered contiguously in RAM. The execution times drop to circa one third in Pandas, half in R and fifth in kdb+. The execution times with the type optimizations on a sorted table of size 1 billion is shown below.
Parallelization
All modern computers have multiple CPU cores. Pandas by default operate on a single core. What do we need to do to make the computation parallel and exploit multiple cores?
Python libraries Dask and Ray are the two most famous libraries for running parallel computations. Library Modin is a wrapper above these engines for Pandas data frames. The "media" is loud from the claim that you gain significant query performance improvements, even on an average laptop, by replacing a single line from

to

This is probably true for a small number of textbook examples but does not apply in real life. With my simple exercise, I run into several issues both with Ray and with Dask. Let me describe the problem one-by-one starting with Ray.
First, the category type is not supported. Second, function apply and agg behaves differently than the corresponding Pandas functions. Using multiple aggregates with a group-by in function agg is not supported so the operation falls back to Pandas operation. The function applies only handles lambdas that return a scalar. This disables the second, more elegant Pandas solution. Furthermore, apply returns a Modin data frame instead of a series. You need to transpose the result and rename the column index (0) to a meaningful column name, e.g.

Finally, the code ran significantly slower than the Pandas equivalent and broke with 100M rows. Pandas and the others handle 1B rows easily.
Moving to the library Dask also has some nuisances. First, it does not handle the weighted average if the bucket’s type is categorical. We lose an important performance improvement technique. Second, you need to provide type hinting to suppress warnings. This means yet another column name repetition. In the more elegant apply-based solution you type the output column names (like TOTAL_QTY) four times ☹️. So it seems moving to Dask is not as simple as extending the code with a simple compute statement to trigger the computation.
Parallelization in kdb+ is automatic for on-disk, partitioned tables and for in-memory tables in version 4.0. You won't observe any type problem - everything works smoothly. All you need is to start kdb+ in multiprocess mode via command line parameter -s. The built-in map-reduce decomposition spreads the computation to several cores for the majority of operations including sum, avg, count, wavg, cor, etc. You can also get performance gain by partitioning the table manually and use function peach that executes functions in parallel. All we need is to change from

to
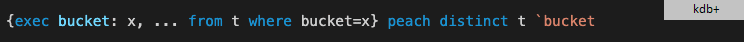
This simple code modification resulted in almost an order of magnitude faster execution on a 16-core box with kdb+ version 3.6. Since version 4.0 already employs parallel computing, manual parallelization adds no value. If you are aiming for the best performance then code will be simpler with kdb+ 4.0 than with 3.6.
R data.table also uses multiple threads by default and executes queries in parallel behind the scene. You can check and set the number of threads that the data.table uses via function setDTthreads.
Let us compare the execution times of the most performant versions of all languages. For SQL we evaluated BigQuery which is considered to be the fastest SQL implementation for huge datasets due to its massive parallelization.
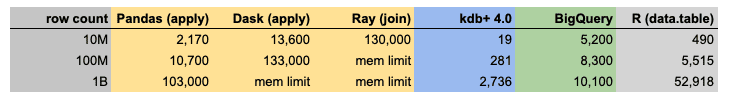
kdb+ is again the winner in this category, BigQuery being the runner-up. R data.table is two times faster than Pandas.
Above 1 billion rows
The largest tables in our experience contained one billion rows. Above this number the table does not fit into the memory, hence Pandas is out of the league. Dask and Ray which is designed for parallel processing and cluster of computers performed poorly compared to the other contenders. For BigQuery the size of the table almost does not matter. The execution time will hardly increase if we move from 1 billion rows to 10 or 100 billion rows.
In kdb+, the data can be persisted to disk so it can cope with terabytes of data. The query will be the same and kdb+ automatically applies map-reduce and leverage multicore processors. Furthermore, if the data is segmented and segments are located on different storages with separate IO channels then IO operation will be executed in parallel. These low-level optimizations allow the kdb+-based solution to scale gracefully.
Distributed computing with kdb+
As a final simple exercise let us investigate how can we spread the computation across many kdb+ processes, leverage our cluster of machines and horizontally partition our sample table. How difficult it is to achieve a Ray/Dask/Spark-like distributed computation with kdb+?
Function peach uses external, slave kdb+ processes as opposed to slave threads if variable .z.pd stores connection handles to slave kdb+ processes.

We can split table t by bucket values

Finally, we can distribute the select statement and merge the result. Function raze is similar to Pandas' concat function. From a list of tables, it produces a large table via concatenation.

Well done! We implemented "kdb-spark" with four lines of code.
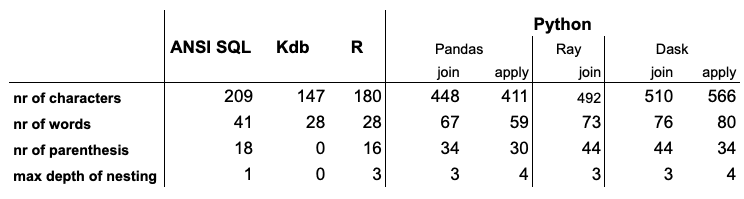
Code simplicity
I collected a few statistics about the code themselves. Although short code does not necessarily mean clean code, for this particular example these metrics well correlate with simplicity.
Conclusion
My observations are collected in the table below. Emoji ????stands for excellent, ✔️for good and ☹️for a disappointing performance.

Python is often the first programming language a student learns. It is simple, performant and has a slight learning curve. Its library Pandas is a natural step to introduce new-joiners to the world of data analyses. Pandas is also often used in a professional environment and more complex data analysis. Pandas looks tempting in simple, textbook exercises but is inconvenient to use in our simple real-world use-case.
There is more focus on extending Pandas and support large tables via cluster computing. Ray, Dask and Modin are in an early phase with a bunch of limitations. In our use case, they simply added syntactical complexity and actually decreased performance.
R outperformed Pandas in every sense, including simplicity, elegance and performance. There are several built-in optimizations, such as inherent multi-threading. Optimization of string representation works like a charm and allows developers to focus on the analyses as opposed to minor technical details. It comes as no surprise that R data.table is being ported to Python. Maybe package data.table will be the replacement of Pandas in the future?
Kdb+ takes data analyses to the next level. It is designed for extreme productivity. In our use case, it was the clear winner of simplicity, elegance, and performance. No wonder that 20 out of the 20 top organizations in the capital market chose kdb+ as a primary tool for data analyses. In this industry, the data analysis drives the revenue and the tools are tested under extreme conditions.
BigQuery is a winner in performance above 100 Billion rows assuming that you don't have a cluster of computers at hand. If you need to analyze huge tables and you are sensitive to run-times then BigQuery does a great job for you.
Related works
DB ops benchmark was initially started by Matt Dowle the creator of R data.table. Besides Pandas, data.table, dask dplyr they also test Apache Spark, ClickHouse and Julia data frames on a variety of queries with different parameters. Anyone can see the queries side-by-side and even download the test environment to conduct the experiments of his/her custom hardware.
Mark Litwintschik uses the NYC taxi drives database and four queries on 33 database-like systems. He provides a detailed description of the test environments, the settings he used and some valuable personal remarks. It is a thorough work that demonstrates Mark's outstanding knowledge in Big Data platforms. His observation is in line with our experiments, kdb+ turned out the be the fastest among the non-GPU-based database solutions.
The STAC-M3 benchmark was originally developed in 2010 by several of the world’s largest banks and trading firms. It is designed to measure exactly how much emerging hardware and software innovations improve the performance of time-series analytics. After STAC-M3 was developed, kdb+ quickly became the favorite database platform for hardware vendors running the tests because it set performance standards that other software providers simply couldn’t beat. Also, Google acknowledged STAC-M3 as an industry-standard benchmark for time-series analytics. They used kdb+ to demonstrate that migrating data and workload from on-premise to GCP does not mean compromising the performance.
Acknowledgments
I would like to thank Péter Györök, Péter Simon Vargha and Gergely Daróczi for their insightful bits of advice.
Bio: Ferenc Bodon Ph.D. is an experienced data engineer, software developer, multi language programmer, software architect with academic background in data mining and statistics. Reflects long-term thinking and always striving to find top quality, robust solutions that are scalable and allow rapid development. Believes in software quality and cannot relax until the "glory solution" is found.
Original. Reposted with permission.
Related:
- 10 Simple Hacks to Speed up Your Data Analysis in Python
- The Last SQL Guide for Data Analysis You’ll Ever Need
- The Berlin Rent Freeze: How many illegal overpriced offers can I find online?

 Python for data analysis… is it really that simple?!?
Python for data analysis… is it really that simple?!?