DeepMind’s AlphaFold & the Protein Folding Problem
Recently, DeepMind's AlphaFold made impressive headway in the protein structure prediction problem. Read this for an overview and explanation.
In case you missed the news, the protein structure prediction problem was solved recently!
A common way to introduce the protein folding problem is by comparing it to something at the macro scale of human experience. It’s almost a trope to compare protein folding to origami, the creative endeavor of making complicated 3-dimensional shapes from relatively uncomplicated 2-dimensional paper sheets. Even xkcd makes the comparison.
In fact it’s a much harder problem than most realize, and exponentially so, because the origami metaphor is missing an entire dimension. It may be difficult to fold an origami crane from a sheet of plain paper, but think about doing the same starting with a piece of floppy string instead.
Let’s keep going with the metaphors:
For CS nerds, it’s like trying to find a minimum description length with O(n) complexity for an O(n3)algorithm, as compared to going from O(n3) to O(n2).
For people that go outside, it’s a bit like the increased difficulty of riding your bike uphill into a strong headwind versus the difficulty of carrying a sandwich in your backpack.
Folding an origami crane from a flat sheet of paper is a common analogy for protein folding, but perhaps a more apt one is folding a paper crane out of string. From left-to-right images are sourced from Ilja Frei on Unsplash and Wikipedia user Laitche, used under the Unsplash License and Public Domain, respectively.
Recently, and to widespread acclaim, critique, and comment , protein structure prediction has been solved!
This constitutes a major advance, and as John Moult was quoted in the MIT Technology Review article, it’s “the first use of AI to solve a serious problem.” At 2020’s Critical Assessment of protein Structure Prediction competition, its 14th iteration (CASP14), DeepMind’s AlphaFold 2 entry achieved a median score of about 92% GDT_TS, a measure of amino acid residues being within an acceptable accuracy threshold of experimentally determined positions across more than 90 structure predictions.
That’s considered by many to be comparable to the experimental accuracy of measuring crystallized protein structure directly, and beats out the next best competition by a significant margin. AlphaFold 2 at CASP14 has been heralded by some as biology’s 2012 ImageNet moment, but that’s almost certainly an understatement.
Despite the significance of the accomplishment and anticipated impacts in both basic and medical research, biology remains far from solved. While an eager young grad student may want to adjust their career trajectory somewhat, biology remains a literal hot mess of complexity and beauty with plenty of confusing mysteries yet to be explored and simulated.
The Problem with Protein Folding
Our first order of business is defining the problem that was so cleverly solved by AlphaFold 2 (in specific circumstances, for particular targets). After the problem is defined, the next stage is differentiating the so-called protein structure prediction problem from the much more difficult and almost entirely unsolved problem of protein folding. More accurate coverage of DeepMind’s placement in CASP14 refers to the solution of a “decades-old grand challenge of biology” or similar rather than “the protein folding problem,” but a substantial number of outlets and even the DeepMind blog post erroneously refer to the latter terminology. In fact, the protein structure prediction problem and the protein folding are quite different in scope, difficulty, and application.
| A basic primer on the central tenets of molecular biology
Molecular biology often makes reference to what's known as the "central dogma." This simplified perspective provides a direct line from a chemical string of letters — DNA — to phenotype, or the actual traits and characteristics of biological organisms.
Of course, it's all a little bit more complicated than that. Actually a lot more. The central dogma is something of an oversimplification of an already oversimplified mental model. In fact every step has myriad ways of modifying itself and interacting with other cellular processes. Much of the non-coding regions that make up the vast majority of animal DNA (previously popularized as “junk DNA”) actually have regulatory functions, either directly as instructions for making interfering RNA, or less directly in modulating transcription from DNA to RNA. Even the final step from an amino acid sequence to a folded protein (the purview of AlphaFold 2) has multiple methods for differential folding and post-translational modification. |
Protein Structure Prediction is No Easy Task
Protein structure prediction is a matter of taking a known amino acid sequence and predicting the associated protein structure. Notably, the predicted structure is compared to a ‘ground truth’ structure determined by experimental procedures such as X-ray (aka Roentgen ray) crystallography, cryogenic electron microscopy, or Nuclear Magnetic Resonance (NMR) spectroscopy.
The vast majority of protein structures available for training a model like AlphaFold 2 are the result of the first method, but protein crystallization is a finicky process and the resulting structures have some caveats. It’s helpful to remember the mantra: “the cytoplasm is not a crystal”, to say nothing of the many other environments proteins naturally operate in within intra and extracellular space.
Protein structure and activity in the wild are modified by small molecules, metal cofactors, cell and organelle membranes, pH, and many other determinants, and they can be quite a bit more dynamic than a static crystal structure.
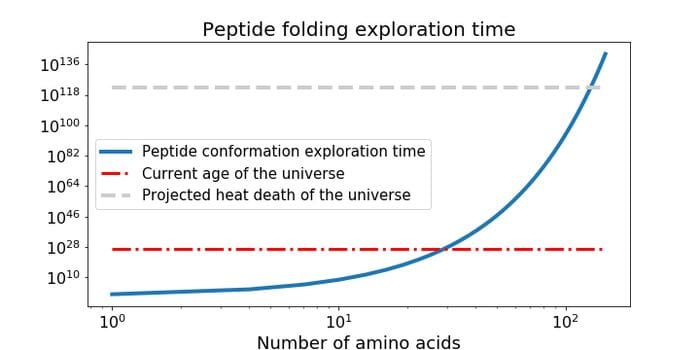
A visual description of Levinthal’s paradox.
Levinthal's Paradox
Any introduction to the protein structure prediction problem would be incomplete without bringing attention to Levinthal's paradox, which emphasizes the intractability of determining protein structures from sequence based on native search alone. The paradox takes its name from the originator of the thought experiment, Cyrus Levinthal, who postulated that it would take longer than the anticipated lifespan of the universe to randomly explore all possible conformations of a protein, even a modestly sized one with only 100 amino acid residues.
The practical lesson here is that even though the compute requirements for training AlphaFold 2 were substantial, and the inference time compute requirement is extraordinary compared to typical norms (more on that later), Sutton's bitter lesson had a diminished role to play, if any, in training AlphaFold 2.
The protein folding problem is another beast entirely, one that is concerned with the dynamic processes a protein undergoes while being translated from a string of RNA nucleotides to an agglomeration of amino acids, ultimately resulting in a fully formed protein structure. Protein folding dynamics depend on local chemistry, temperature, and pH but can also be heavily modified by heat shock proteins and other molecular chaperones.
This problem remains untouched by DeepMind’s AlphaFold project for now.
Focusing on a Sub-Problem
Within the protein structure prediction problem the team at DeepMind has focused on a very specific sub-problem: that of structure prediction for proteins composed of a single amino acid chain. In fact, many proteins operate in the cell as complexes, which can vary in complexity from a pair of the same protein (i.e. dimers) to massive complexes of some 10s of different proteins. These protein structures in turn may interact with many different molecules in the cell in a dynamic fashion.
Unsurprisingly, many critical reviews of AlphaFold 2 tend to stick to the ‘protein folding’ label, which is perfectly fair so long as DeepMind insists on using the same in their own public relations efforts. But we’re not here to make this a semantic argument; instead we're concerned with exactly what was accomplished. We can qualify the accomplishment as the solution of the single-chain (mostly crystallized) protein structure prediction problem for proteins amenable to measuring their experimental structure. Even with a hefty string of qualifiers in front of it, AlphaFold 2’s achievement is likely to have significant impacts in a number of areas of applied and fundamental science over the next few years.
The Achievement by the AlphaFold Team at CASP14
In the last section, we clearly delineated the bounds of the problem solved by the AlphaFold team at CASP14. This is something that is sorely missing from much of the lay-press and public relations coverage of the event, and consequently forms the basis for most of the contention from skeptics.
While AlphaFold 2 did not solve, nor did it attempt to, the protein folding problem per se, it does seem to have solved a very specific version of the protein structure prediction problem, defined as achieving a GDT_TS score of 90 or above for single-chain proteins in the biennial CASP competition.
In this section we’ll briefly discuss the protein targets used to evaluate CASP14 and the various scoring metrics used. This information should help protein structure enthusiasts of all kinds to better calibrate their understanding of CASP14 results and put the huge gap between the performance of AlphaFold 2 and the next tier of competitors into perspective.
These competition details are not strictly necessary to get a feel for the broader impact, and machine learning enthusiasts eager to geek out on (mostly speculative) details about the AlphaFold 2 model may be more interested in reading the next section first.
The Most Reported Metric is GDT_TS
The most reported metric describing CASP results is GDT_TS. The “GDT'' in “GDT_TS” stands for global distance threshold, and “_TS” designates that the metric is an average of GDT for 4 different thresholds, namely 1, 2, 4, and 8 Angstroms. For example, with 100% of residues within 4 (and thus 8) Angstroms, 90% within 2 Angstroms, and 80% within 1 Angstrom would give a GDT_TS of 92.5, which is pretty close to the overall GDT_TS reported for AlphaFold 2. The curve for target T1038 from CASP14 gives a pretty good idea of the gap between AlphaFold 2 and the competition, albeit not all targets had such stark a difference.
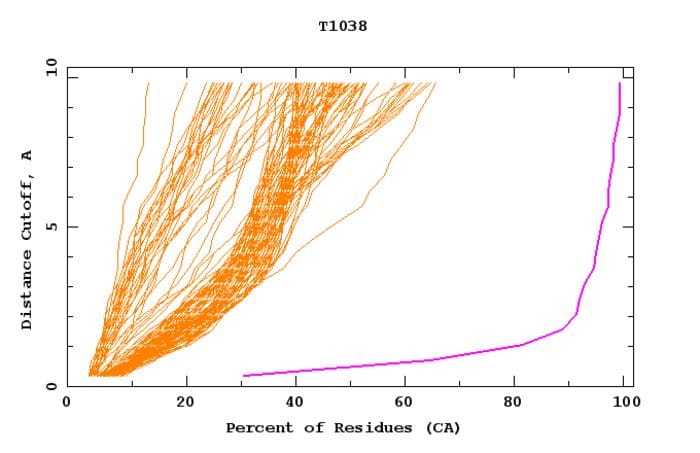
Global distance threshold (GDT) curve for CASP14 target T1038. The curve for AlphaFold 2 is shown in purple. Credit to CASP.
Another metric featured prominently on CASP leaderboards is the Z-score. Protein folding can be thought of as the biophysical process of amino acid chains finding the lowest energy conformation, similar to how a spring would rather be uncompressed, water prefers to find the ocean, or oil droplets would rather separate themselves from water. Protein structure prediction is likewise trying to find this low-energy state given a protein sequence, and the Z-score is an estimate of the deviation in total energy of the protein structure relative to the energy distribution of the protein sequence with random folds.
CASP does do some processing and outlier removal to encourage novel approaches, but for the numbers you see reported on CASP leaderboards, higher is better. A full discussion of Z-scores and how they are calculated is beyond the scope of this article, but for the curious a good place to start would be Jacob Stern’s series on Medium.
The Model
Unfortunately we’re still waiting for a full description of AlphaFold 2. The wait between the CASP13 results released at the end of 2018 and the AlphaFold paper published in Nature at the beginning of 2020 was substantial. That huge delay meant that trRosetta, a deep learning + energy minimization model for predicting protein structures (in many respects an open source version of the original AlphaFold) was published in PNAS just 6 days after AlphaFold was finally described in Nature (and the MIT-licensed GitHub repository was put up several months earlier). Here’s hoping it doesn’t take as long this time for DeepMind to make their work public.
We can speculate about the details of AlphaFold 2 based on what DeepMind told us in their blog post and what CASP14 attendees were able to glean from the paltry presentation given by the AF2 team. The model description below is somewhat speculative, but should capture the ideas underlying DeepMind’s CASP14 success at a high level of abstraction.
Unlike last time at CASP13, AlphaFold 2 is an end-to-end differential system of several neural network modules.
The first stage contains an embedding model and neural network that predicts pairwise distances between amino acid residues. That much is similar to the first AlphaFold, except instead of applying constrained gradient descent over the model output to estimate protein structure, features learned by this model and another also contained in the first stage are given directly to a structure model. We can infer from the similarity to the first AlphaFold that this module is probably a residual convolutional neural network with dilated convolution kernels, and we’ll refer to it as NN0.
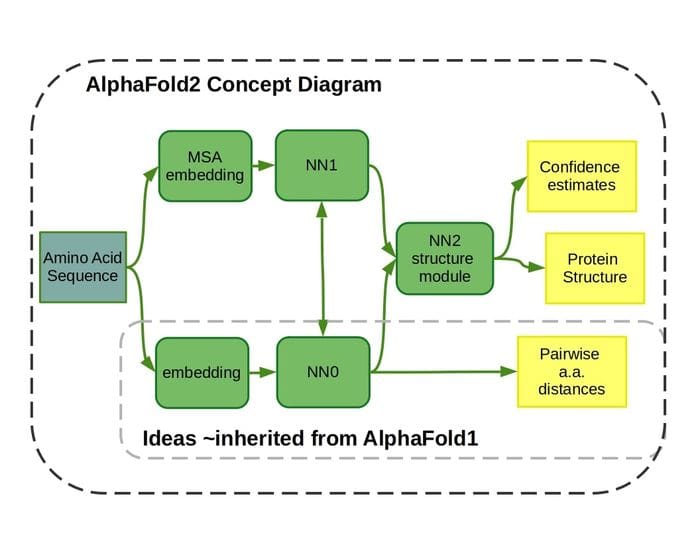
High-level overview of AlphaFold 2.
The other branch in the first stage of AlphaFold 2, which we’ll refer to as NN1, is almost certainly some sort of graph neural network, based on descriptions from DeepMind and others. The inputs to this neural network module are a collection of multiple sequence alignments, i.e. sequence fragments that appear evolutionarily related to the sequence in question, and for which science knows something about the structure. The model uses an attention mechanism to iteratively focus on different combinations of MSA inputs. It’s not clear if the number of iterations is determined beforehand.
We suspect that iterations continue until the overall structure prediction surpasses a confidence threshold, or some maximum number of several hundred iterations is reached. The inclusion of a confidence estimate at the output and the significant computational cost at inference time supports the idea of the model itself determining when it has converged on an acceptable solution.
Not a whole lot is known about the third module (we’ll call it NN2) that receives information from the first stage. It’s described in the DeepMind blog post as a “structure module” and Professor Mohammed AlQuraishi of Columbia University supposes that it is probably some variant of a transformer with rotational invariance, i.e. an SE(3) Transformer.
Controversy with Rotational Invariance
Rotational invariance (or equivariance) is particularly important for understanding chemical structures, but it’s also been a bit of a thorn in the side of more traditional image-based deep learning.
A crash in 2020 involving Tesla’s autopilot seems to have been at least in part due to a failure to recognize an overturned truck as an obstacle. While convolutional neural networks of the sort used in image recognition for autonomous vehicles famously exhibit translational invariance (an important factor behind their many successes), they fall short when presented with objects in orientations not seen during training.
In image-based conv-nets, data augmentation can be used to expose models to different object distortions like shear and rotation. This helps models to recognize objects in unusual orientations, but it’s a far cry from achieving rotational invariance. It will be interesting to learn how AlphaFold 2 incorporated rotational invariance in NN2, when the inevitable top-tier publication is released. Whether it’s essentially an SE(3) transformer, borrowed from published work such as tensor field networks, or does something entirely different remains to be seen.
We can’t end this section without pointing out that the lack of details coming from DeepMind about their seminal performance at CASP14 is a substantial disappointment compared to their community participation at CASP13. According to Mohammed AlQuraishi, DeepMind’s CASP14 presentation was “almost entirely devoid of detail” and “barely resembled a method talk” at all. This is part of a larger trend evinced by high profile and well-resourced commercial AI labs.
Should we expect AlphaFold 2 to be hidden behind an API, like GPT-3, to be licensed at great expense mainly to the likes of commercial pharmaceutical companies? Time will tell, but if DeepMind is going to rely on massively international and largely publicly funded resources like the Protein Data Bank (PDB) for training, and the CASP experiment for validation, hopefully they will return the favor by providing sufficient information to replicate AlphaFold 2 with all due haste.
The Impact of AlphaFold’s CASP14
While there remains substantial skepticism pending the release of details (and ideally code!) for AlphaFold 2, researchers are already showing enthusiasm for what AlphaFold 2 can do for protein structure science and biology as a whole.
Some say AlphaFold 2 may do for structural proteomics what DNA sequencing did for genomics. As reported in Science, Andrei Lupas of the Max Planck Institute for Developmental Biology and CASP assessor, has already used AlphaFold 2 to find the structure of a particularly tricky membrane protein.
DeepMind has signaled that the next problem the AlphaFold team will try to solve is structures for multi-chain protein complexes. This is probably not such a giant leap from what they’ve already done though, and other groups given access to AlphaFold 2 would probably be able to make substantial progress here as well.
Protein design for pharmaceuticals is another area they seem to be interested in, and of course that one sounds like it could be quite profitable.
As we saw after DeepMind entered the field of protein structure prediction in CASP13, when prominent commentators wondered why well-resourced pharmaceutical labs and decades of efforts by academics had been so thoroughly scooped, the field is facing a reckoning of sorts. The gap between AlphaFold 2 and the competition was substantial, and in fact the gap between 2nd tier entries from the Baker Lab, which used a lot of ideas from the first AlphaFold, and the rest of the field was also quite stark. This may leave some reconsidering their chosen field or, for a young computational scientist, whether or not they should study protein structure in the first place.
The best advice on how to adapt basic research in the fields of biomolecular machine learning and computational biochemistry (and how this might affect one’s own career) comes from the blog of Professor AlQuraishi mentioned earlier. Read the last section of that post for a nuanced perspective. In general, his advice boils down to focusing on the open-ended side of research, like which questions are worth answering, and not focusing on already defined problems with clear metrics for success (like the CASP leaderboard) and plentiful training data that could fuel the next AlphaFold.
Instead look to areas like protein-ligand interaction and in situ molecular dynamics. After all, AlphaFold 2 may have largely solved the structure-from-sequence prediction problem for proteins in crystal form, but the cytoplasm, where all the chemistry of life takes place, is most assuredly not a crystal.
Concluding Thoughts on AlphaFold 2
In a field where large research groups make announcements about surpassing human performance for a new task on a near-monthly basis, AlphaFold 2 clearly stands out. Not only is it a major breakthrough in the protein structure prediction problem, but it showed what a clever model can do in a task that is basically intractable for a human without computer assistance.
We’ll all be eagerly awaiting more details to understand the reason behind an anomalously high compute budget at inference time, a major departure from the usual case where a neural network might provide a shortcut for a more intensive physical simulation.
Neural networks have definitely been shown to be capable of approximating other tasks in science, but these applications rarely get used beyond the proof-of-concept stage and can draw heavy criticism about the narrow application space.
AlphaFold 2 might be very different, with CASP assessor Andrei Lupas reporting that AlphaFold 2 has already helped his lab to solve a structure they’ve been working on for 10 years. The most prominent barrier for similar impacts to be widespread will be how DeepMind handles the rollout of the work. Will they try to keep the details somewhat proprietary and hidden behind a license, or will they embrace the open nature of science and ensure everyone has access to the relevant details? If they stick to the former strategy you can expect they may get leap-frogged by an open source alternative that shares most of AlphaFold 2’s capabilities sometime in the next year or so.
AlphaFold 2 is also a pretty good example of what AI can do that goes beyond "human-level" intelligence, instead representing a sort of hybrid of the types of problem-solving and creativity that humans are good at with those that computation excels at.
Usually, it’s a lot harder to train machines to do the things that humans take for granted, while the types of things that impress other humans at parties (chess, trivia, rapid numerical calculation) are comparatively easy for machines. From a human perspective we might think of those domains as usually being at odds with each other, but the synergy of the two may ultimately be the source of the most surprising results and capabilities of eventual artificial general intelligence.
Original. Reposted with permission.
Related:
- Vision Transformers: Natural Language Processing (NLP) Increases Efficiency and Model Generality
- GPT-2 vs GPT-3: The OpenAI Showdown
- Compute Goes Brrr: Revisiting Sutton’s Bitter Lesson for AI


